Skeleton thickness and porosity_fiji
#Skeleton thickness and porosity_Fiji
#Skeleton thickness and porosity_Fiji
#Respirometer Manual
Written by Maggie Schedl
This protocol details the step-by-step use of the PreSens/Loligo Microplate respiration system for measuring respiration and photosynthesis of coral planulae.
Using the EpoFix Resin and following the EMS instructions for the preparation of the resin.
Ten days old spats of the coral Stylophora pistillata were fixed with resin for synchrotron radiation imaging (BAMline) at BESSY II in Berlin.

10 ml ampoules 16% Formaldehyde -EMS Sciences Cat #15710 Sodium Cacodylate Trihydrate bioxtra10 g - Sigma Cat #C4945 Na2CO3 Epofix
22 g/L Na2CO3 in water filtered 0.2um
0.05M Na Cacodylate Buffer in 22g/L Na2CO3 Add 10.7 gms Cacodylic acid (to 1000 ml 22g/L Na2CO3)
2% Formaldehyde in Sodium Cacodylate Buffered Sea-Water Fixative Add 10 ml ampoule of 16% Formaldehyde to 70 ml of the made up buffer
Laminin (Sigma # L 2020, 1mg/ml) Aliquot into 20μl units Store at -20ºC • Add diluted Laminin solution to cell suspension at a ratio of 1 part Laminin solution to 3 parts cell suspension.
7.6 g Na2B4O7 (Sigma # 221732) 200ml DDW pH 8.2
10mg Poly-D -lysine (Sigma# P 1024) 10ml Sodium borate solution Filter Sodium borate solution only Keep for 30 minutes at room temperature, Store at -4º
2.375g Na2B4O7*10 H2O (Sodium Tetraborate) (Sigma # S 9640)
1.550g H3BO3 (Boric Acid) (Sigma # B 6768)
500ml DDW
pH-8.5 with 1M HCL
Filter and store at 4◦C
0.1ml or 0.107g 50% PEI (Sigma# P 3143, 50ml)
49.9ml Borate Buffer
Aliquot into 1ml amounts,
Store at -70◦C
Before using : thaw 1ml aliquots and add 9ml Borate Buffer
Store rest at -20◦ C for no more than 1 mounth.
Use about 50μl/cm2 Poly-D-lysine / Poly-Ethylene-Immine solution
Coat overnight at room temperature,
Aspirate solution using sterile Pasteur pipette
Wash in DDW,
Dry with sterile Pasteur pipettes.
Using the DMEM no glucose and antibiotic-antimycotics
Add the follow to 1l of deionized H2O and filter with 0.2µm keep stock in 4C and worm to 25C before using
| chemical comment | gram |
|---|---|
| NaCl | 23g |
| KCL | 0.763g |
| MgSO4-7H2O | 1.89g |
| MgCl2-6H2O | 10.45g |
| Na2SO4 | 3g |
| NaHCO3 | 0.25g |
| SrCl2 | 0.026g |
Add to [DMEM no glucose] (https://www.thermofisher.com/il/en/home/technical-resources/media-formulation.49.html) the follow
| chemical comment | gram | comment |
|---|---|---|
| NaCl | 9.05g | |
| KCL | 0.7g | |
| CaCl2 -2H2O | 0.71g | |
| MgCl2-6H2O | 5.1g | |
| Na2SO4 | 0.5g | |
| taurine | 0.5m | from stock 1.04µg/20ml DDW |
| NaOH3- | 1.85g | only if necessary, if the DMEM bought is without NaOH3 |
| Hepes | 2.98g |
Keep stock in -20C
Dilute the stock DMEM culture medium to 12.5%(vol/vol) in ASW: 20ml DMEM + 140ml ASW
Add to the diluted DMEM mix :
All work must be done in the biological/laminar flow hood with filtered medium All tips should be stored in the hood. Tips, pipettes, etc. should be exposed to UV light for at least 10 minutes
All medium need to be in 26C
Work with 6wells or 5ml petri-dish
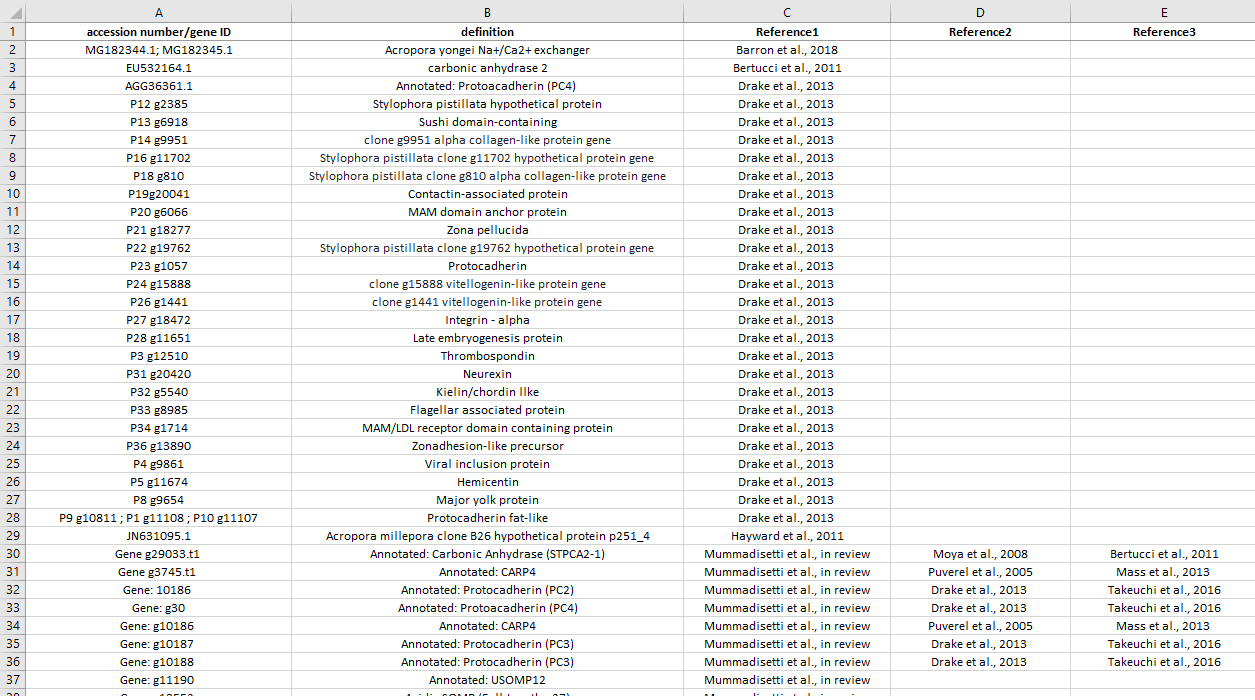
This list comprehends all biomineralziation-related genes known from literature. The first reference is the one related to the accession number/gene ID, then we put all other references where that specific gene/protein was previously detected/immunolocalized (chronological order).
List preview:
These instructions allow to quantitatively assess coral physiological parameters, i.e. tissue biomass (protein concentration), algal density and chlorophyll concentration.
These instructions cover two ways to get algal cell counts using Fiji, one manual and one automated. A fluorescence microscope (Nikon Eclipse Ti, Japan) was used to image symbiotic algae (isolated from coral tissue) both in brightfield and in fluorescent light using 440 nm emission, to identify chlorophyll and to ensure counting of symbiont cells only.